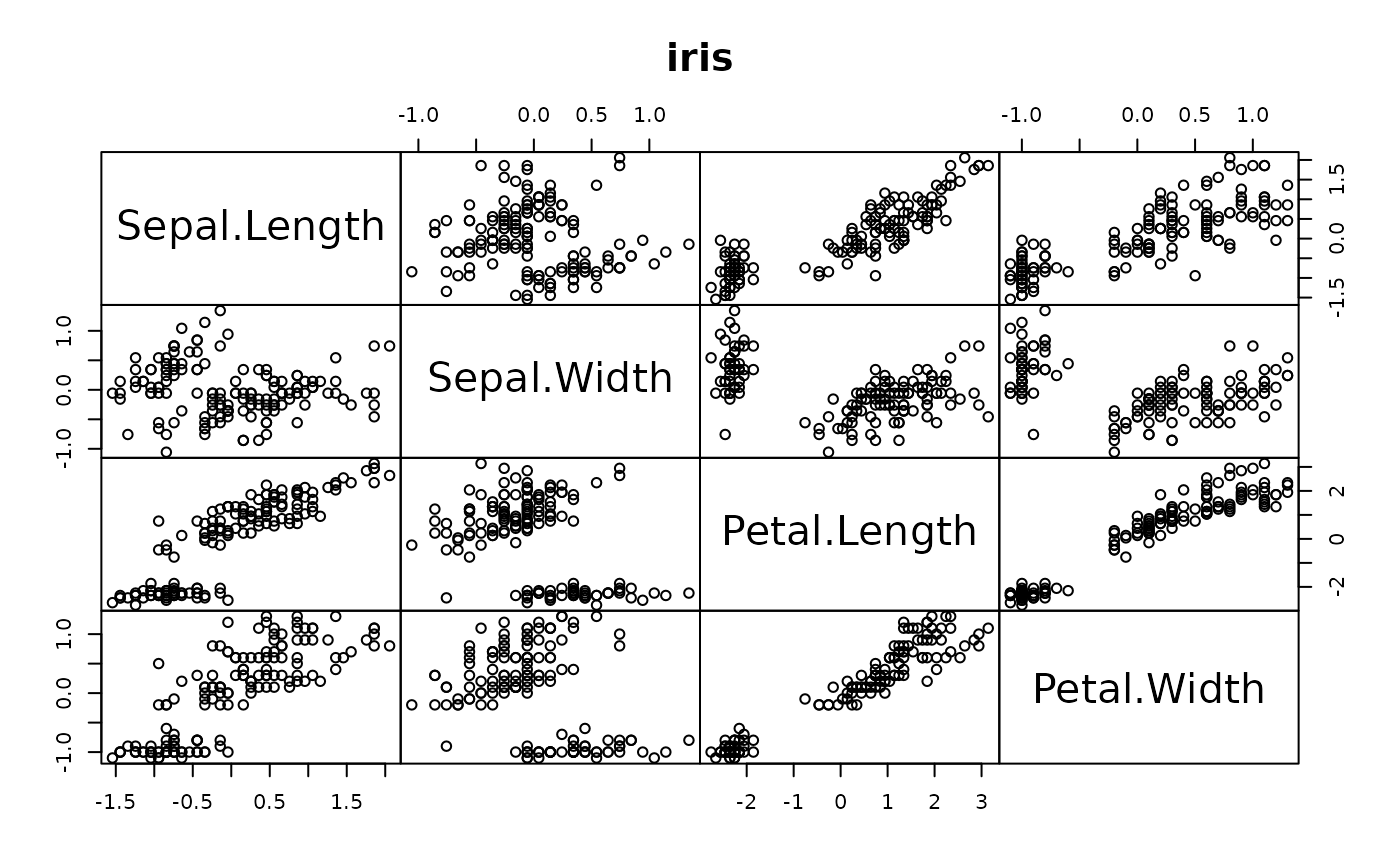
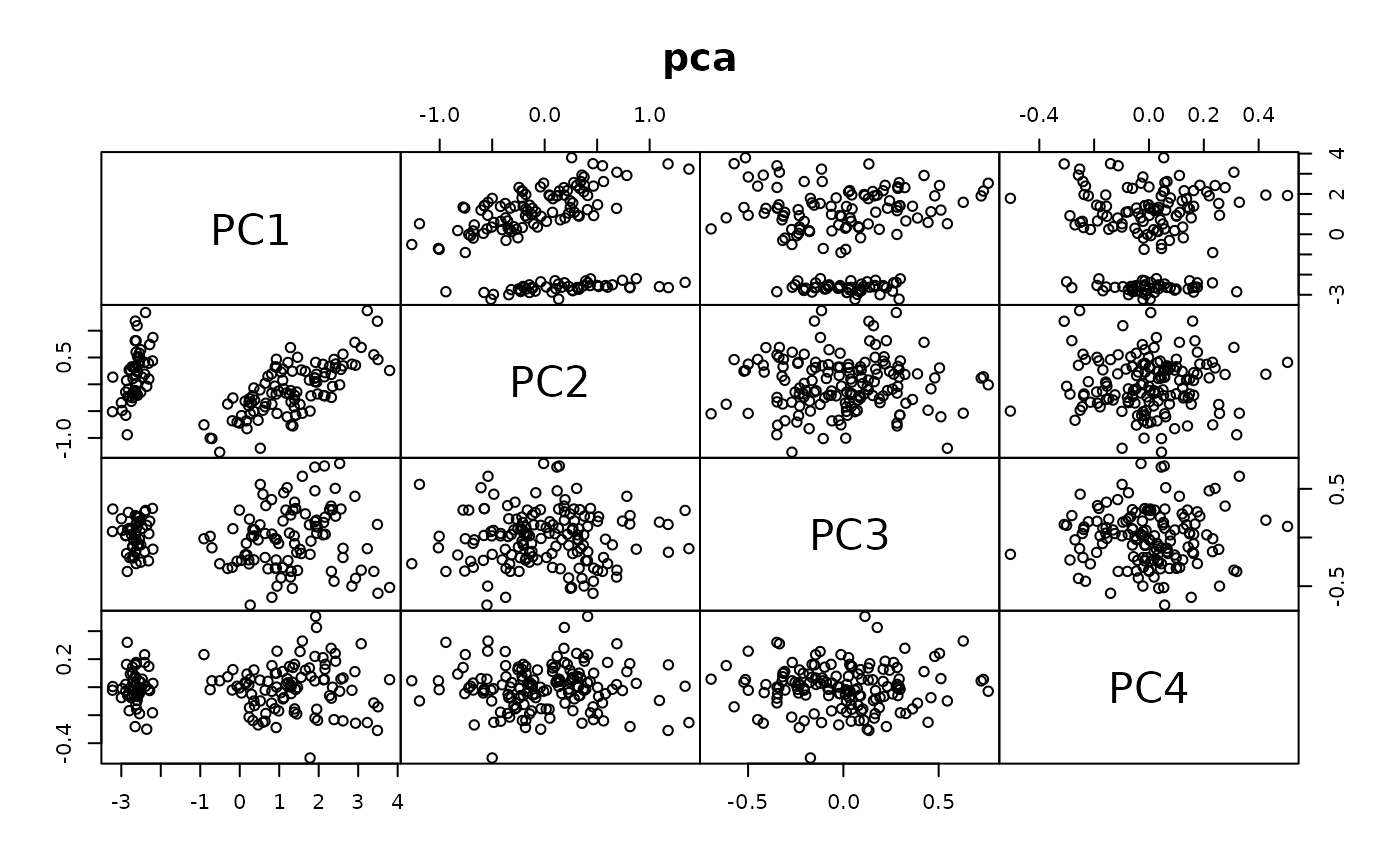
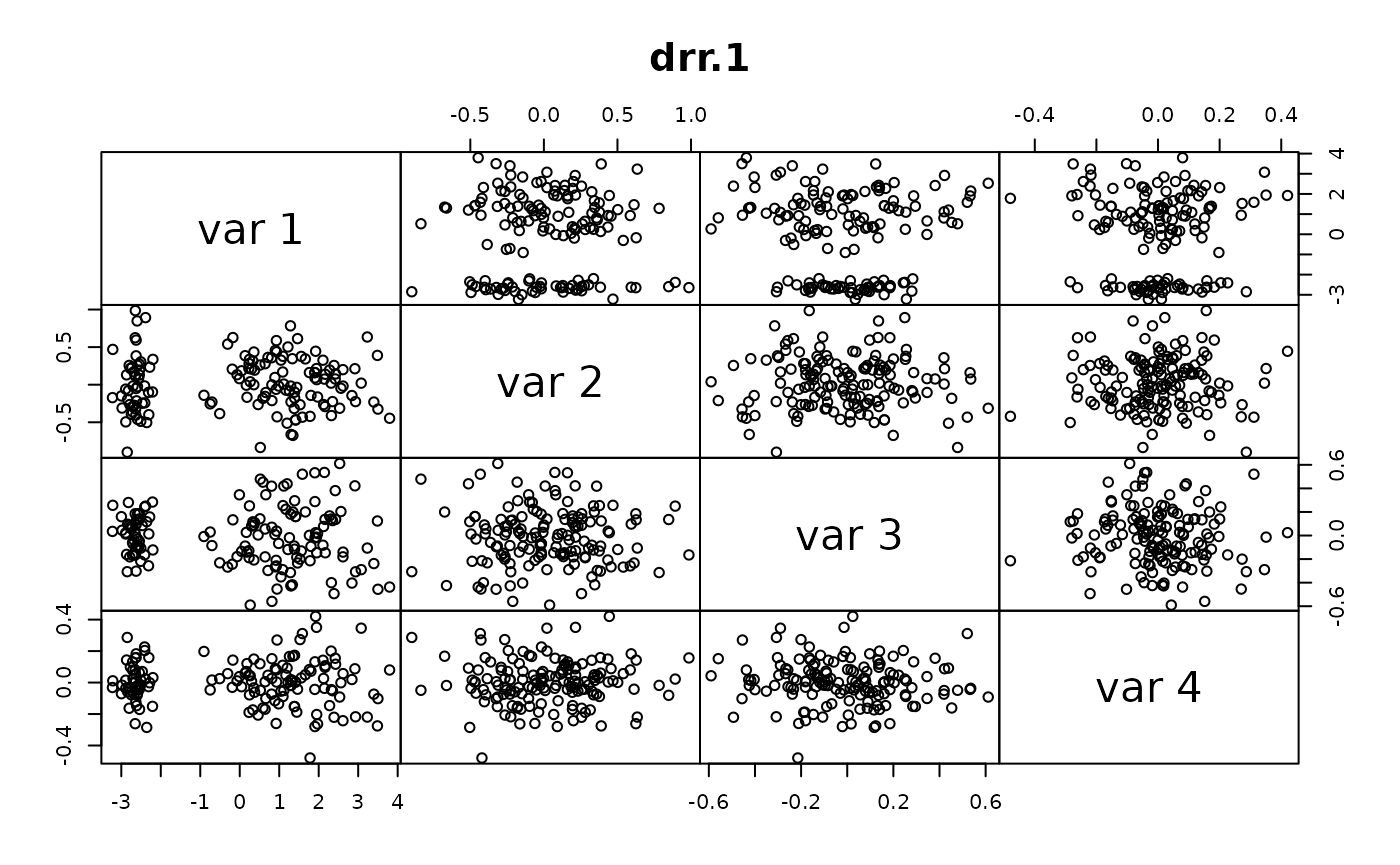
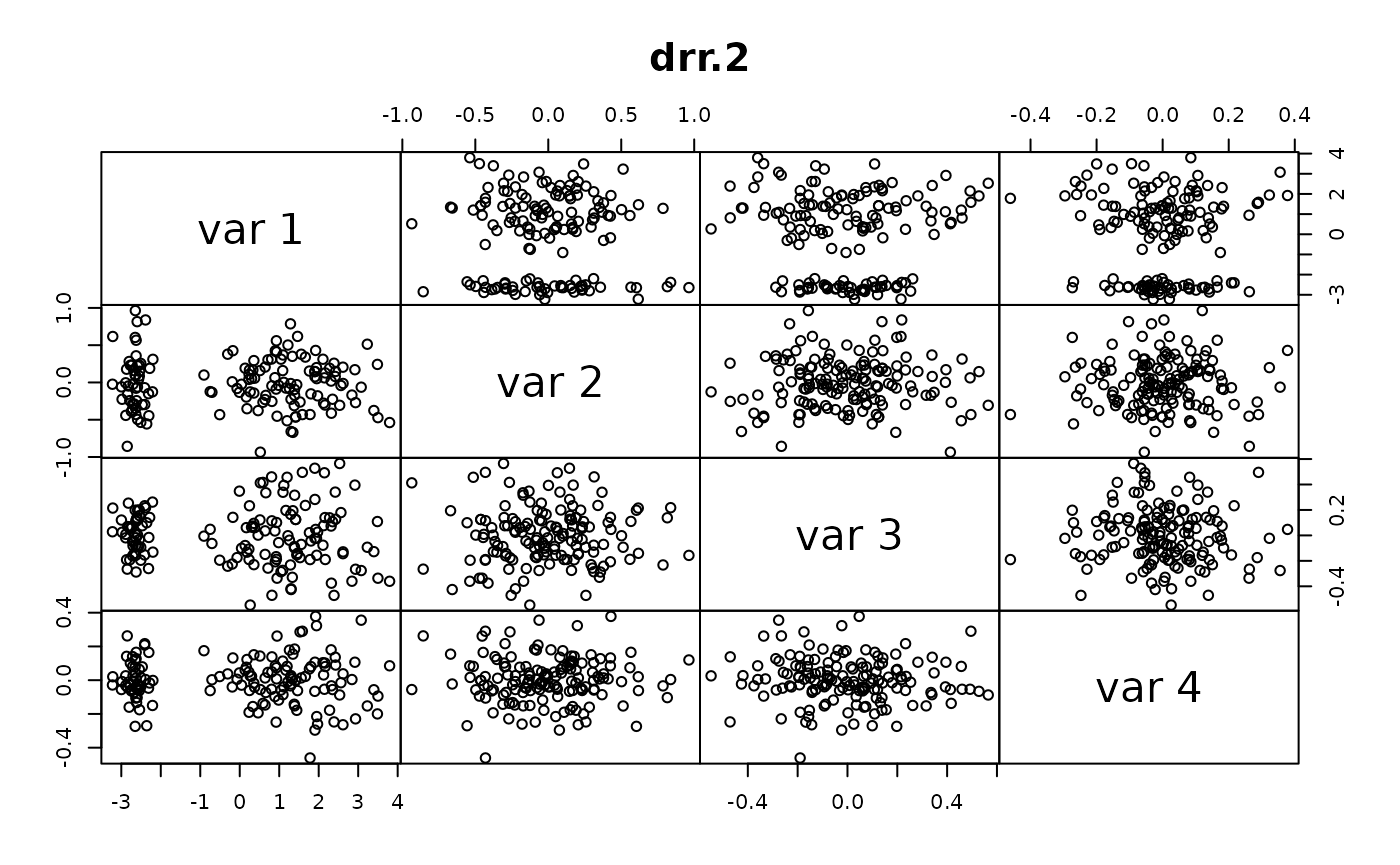
This is an example application to compare the accuracy and computational speed of DRR for different parameters to PCA.
Fit the dimensionality reductions.
t0 <- system.time(pca <- prcomp(my_data, center = FALSE, scale. = FALSE))
t1 <- system.time(drr.1 <- drr(my_data, verbose = FALSE))
t2 <- system.time(drr.2 <- drr(my_data, fastkrr = 2, verbose = FALSE))
t3 <- system.time(drr.3 <- drr(my_data, fastkrr = 5, verbose = FALSE))
t4 <- system.time(drr.4 <- drr(my_data, fastkrr = 2, fastcv = TRUE,
verbose = FALSE))Calculate RMSE
rmse <- matrix(NA_real_, nrow = 5, ncol = nvars,
dimnames = list(c("pca", "drr.1", "drr.2", "drr.3", "drr.4"),
seq_len(nvars)))
for (i in seq_len(nvars)){
pca_inv <-
pca$x[, 1:i, drop = FALSE] %*%
t(pca$rotation[, 1:i, drop = FALSE])
rmse["pca", i] <-
sqrt( sum( (
my_data - pca_inv
) ^ 2 ) )
rmse["drr.1", i] <-
sqrt( sum( (
my_data - drr.1$inverse(drr.1$fitted.data[, 1:i, drop = FALSE])
) ^ 2 ) )
rmse["drr.2", i] <-
sqrt( sum( (
my_data - drr.2$inverse(drr.2$fitted.data[, 1:i, drop = FALSE])
) ^ 2) )
rmse["drr.3", i] <-
sqrt( sum( (
my_data - drr.3$inverse(drr.3$fitted.data[, 1:i, drop = FALSE])
) ^ 2) )
rmse["drr.4", i] <-
sqrt( sum( (
my_data - drr.4$inverse(drr.4$fitted.data[, 1:i, drop = FALSE])
) ^ 2) )
}The Results
More blocks for fastkrr speed up calculation, too are bad for accuracy.